Введение. Инфекции мочевыводящих путей (ИМВП) относятся к наиболее распространенным заболеваниям как в амбулаторной, так и в госпитальной практике среди населения всех возрастных групп во всем мире [1]. Этиологическая структура ИМВП определяется ведущей ролью эшерихий: уропатогенные штаммы Escherichia coli (UPEC) составляют до 90% бактериальных культур при неосложненных ИМВП у пациентов поликлиник и 30–50% – у людей, находящихся на стационарном лечении [2, 3]. В последнем случае воспалительный процесс в органах мочевыделительной системы связан с инструментальными вмешательствами, использованием катетеров и часто вызван полимикробными ассоциациями микроорганизмов [4, 5].
Представители UPEC имеют ряд характеристик, обусловливающих адаптацию бактерий в мочевыводящих путях и последовательную реализацию уропатогенного потенциала на всех этапах инфекционного процесса [2, 6]. Основным механизмом сохранения UPEC в условиях биотопа является их высокая способность к адгезии и биопленкообразованию. Широкое применение в урологической практике био- и искусственных материалов, на поверхности которых формируются бактериальные биопленки, повышает риск персистенции возбудителя в организме хозяина [7]. Среди UPEC преобладают бактерии филогенетической группы B2 (80–85%), около 15% штаммов принадлежат к филогруппе D [8, 9]. Именно у представителей данных филогрупп по сравнению с другими E. coli чаще детектируют гены, кодирующие факторы патогенности, опосредующие цитотоксическое действие, адгезию клеток и т.п., а также обнаруживают маркеры антибиотикоустойчивости [10–12]. В последние годы появляются данные о расширении филогенетического разнообразия UPEC, и возбудителями ИМВП все чаще становятся штаммы E. coli «неуринарных» филогрупп [13].
Представления о популяционной изменчивости UPEC, а также о механизмах адаптации бактерий к условиям урологического тракта базируются на регулярном скрининге биологических свойств возбудителя, проводимом с учетом типа медицинской организации, нозологических форм ИМВП, гендерных и возрастных особенностей пациентов. Активно изучаются патогенные свойства штаммов E. coli, выделенных при осложненных, в том числе катетеризацией, формах заболеваний, а также постоянно контролируется устойчивость микроорганизмов к антибиотическим препаратам и ее корреляция с вирулентностью.
Цель исследования: изучить биологические свойства и филогенетическое разнообразие штаммов E. coli, выделенных при ИМВП у амбулаторных и стационарных пациентов.
Материалы и методы. Объектами изучения служили клинические культуры E. coli, изолированные из материала (моча, катетеры) пациентов с ИМВП, находившихся на амбулаторном (7 медицинских организаций) или стационарном (9 медицинских организаций, 13 отделений) лечении в г. Перми за 2017 г. Всего было изолировано 198 штаммов, из которых 105 обозначены как поликлинические и 93 – как нозокомиальные (выделены через 48 ч после госпитализации).
Определение чувствительности штаммов к антибактериальным препаратам проводили согласно клиническим рекомендациям «Определение чувствительности микроорганизмов к антимикробным препаратам» Межрегиональной ассоциации по клинической микробиологии и антимикробной химиотерапии (МАКМАХ, версия-2015-02). Продукцию бета-лактамаз расширенного спектра (БЛРС, ESBL) детектировали с помощью метода «двойных дисков» согласно методическим указаниям МУК 4.2.1890-04. Гемолитическую активность выделенных культур оценивали на 5%-ном кровяном агаре по диаметру зоны гемолиза вокруг колонии в мм. Биопленкообразование изучали согласно G. А. O'Toole et al. [14].
Выделение тотальной ДНК чистых культур проводили по методике, описанной G.G. Stone et al. (1994): отдельную колонию каждого штамма ресуспендировали в 0,5 мл сверхчистой воды в пробирках типа Эппендорф, инкубировали в твердотельном термостате «Термит» (Россия) в течение 10 мин при 98°C, охлаждали и центрифугировали 5 мин при 13 тыс. об./мин. Надосадочную жидкость использовали для генетических исследований немедленно или после хранения при -18°С.
Филогенетические группы UPEC определяли методом ПЦР (quadriplex PCR) согласно О. Clermont et al. (2013), используя специфические маркеры филогенетических групп и ключ для их установления [15]. Протоколы амплификации соответствовали рекомендациям авторов. Олигонуклеотидные праймеры были синтезированы ООО «Синтол» (Москва). Амплификацию проводили на термоциклере DNA Engine Dyad Thermal Cycler («Bio-Rad», США). Визуализацию полос и документирование данных осуществляли с использованием системы гельдокументации Gel-Doc XR («Bio-Rad», США).
Статистическую обработку полученных данных проводили с использованием компьютерных программ Microsoft Excel 2016 и STATISTICA 10. Для выявления статистически значимых различий двух независимых выборок использовали критерий χ2, критерий χ² с поправкой Йейтса или точный критерий Фишера.
Результаты. В годичном исследовании культуры E. coli изолировали чаще от женщин и в основном от взрослых пациентов лечебных учреждений (табл. 1). Показатель бактериурии ≥103 КОЕ/мл выявлен более чем в 85% случаев и достоверно не различался в материале поликлиник и стационаров. При сравнении образцов, взятых у мужчин и женщин, взрослых и детей, разницы в обсемененности материала не обнаружено. Поскольку нозокомиальные UPEC были выделены как из мочи – уринарные штаммы, так и с поверхности катетера – катетер-ассоциированные штаммы, дополнительно проведен сравнительный анализ этих групп по всем изучаемым свойствам.

В монокультуре UPEC высевались из мочи пациентов с ИМВП в 63,6% (n=126) случаев, тогда как в ассоциации – в 36,4% (n=72). Однако если в поликлинической группе доля ассоциаций составила только 22,9%, то в нозокомиальной – 51,6% (р<0,0001; табл. 2), при этом двухкомпонентные ассоциации встречались в подавляющем большинстве в обоих вариантах. В условиях стационара уринарные штаммы E. coli в составе ассоциаций выделялись в 43,8% случаев, тогда как катетер-ассоциированные – в 80% (p<0,01). Преобладающими (78,1%; p<0,01) при неосложненных ИМВП были двухкомпонентные ассоциации, при катетеризации двух-, трех- и четырехкомпонентные ассоциации встречались одинаково часто, и последних было существенно больше, чем без катетеризации (p<0,005). Доля ассоциантов с микробным числом ≥105 КОЕ/мл была достоверно выше в смешанной культуре с нозокомиальными штаммами UPEC, чем с поликлиническими (57,5 против 37,5%; p<0,05), при этом сопутствующие бактерии чаще выделялись в этой концентрации у катетеризированных больных (90,9%, р<0,01) по сравнению с группой без катетеризации (данные не представлены).
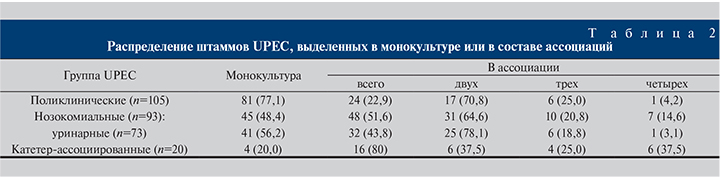
Анализ таксономического состава ассоциаций выявил, что разнообразие видов и их встречаемость существенно различались в двух исследуемых группах. Энтерококк доминировал в ассоциациях как с поликлиническими, так и с нозокомиальными UPEC (58,3 и 54,2% соответственно). В 9 случаях и в той, и в другой группе вместе с E. coli были обнаружены клебсиеллы (37,5 против 18,8% ассоциаций). Следующими по значимости оказались представители рода Corynebacterium (20,8 против 14,6%). Среди госпитальных ассоциаций встречались протеи, морганеллы, неферментирующие грамотрицательные бактерии – псевдомонады и ацинетобактеры, стрептококки и кандиды, не обнаруженные в материале амбулаторных больных. Сравнение нозокомиальных культур, выделенных при ИМВП из мочи и с поверхности катетера, показало, что при катетеризации видовой спектр ассоциантов ограничен несколькими видами: клебсиелла, протей и неферментирующие бактерии встречались достоверно чаще (р<0,005), тогда как при неосложненных ИМВП изолированы коринебактерии, стрептококки, кандиды, которые вообще не были обнаружены при катетеризации (рис. 1).
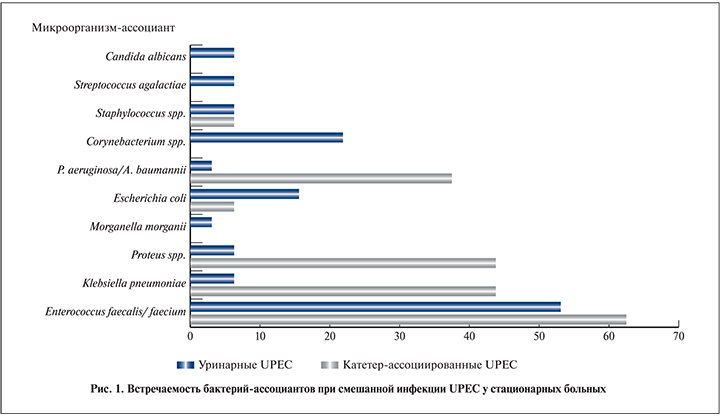
Использование quadriplex ПЦР позволило определить филогенетическую группу более чем у 85% штаммов UPEC (рис. 2, табл. 3). Среди поликлинических культур обнаружены представители всех филогрупп: чаще всего встречались штаммы E. coli филогрупп B2 (37,1%), Е (13,3%) и F (8,6%). Нозокомиальные культуры почти в 90% случаев принадлежали к филогруппе B2, к которой отнесены и все катетер-ассоциированные штаммы. Выявлено, что E. coli филогруппы B2 как в моновидовом варианте, так и в полимикробных ассоциациях достоверно чаще встречаются в стационаре, чем в поликлинике (p<0,00001), тогда как бактерии филогруппы Е, наоборот, – в поликлинике (p<0,0001). Большая часть штаммов E. coli, филогенетическую принадлежность которых не удалось установить данным методом, была изолирована при амбулаторных ИМВП, в стационаре таких культур оказалось только четыре (p<0,001). Оценка гемолитической активности UPEC не выявила разницы в частоте встречаемости данного признака у поликлинических и нозокомиальных штаммов. Не удалось установить связи между наличием гемолиза и принадлежностью к филогруппе В2 в общей популяции UPEC, при этом в стационаре гемолитические E. coli филогруппы В2 встречались достоверно чаще, чем в поликлинике (р<0,001).
Анализ биопленкообразующей способности штаммов UPEC показал, что степень проявления данного признака варьировалась в широких пределах: от 0,054 до 0,593 ЕД. ОП570 (Ме [Q1–Q3] составили 0,132 [0,101–0,188]). Распределение UPEC по степени биопленкообразования выявило, что поликлинические культуры не формировали биопленку в 24,8% случаев, стационарные – в 34,4%, при этом в последней группе небиопленкообразующими чаще были штаммы с катетеров, чем уринарные культуры. Связи между биопленкообразованием и принадлежностью к филогруппе В2 не выявлено ни в общей, ни в отдельно сравниваемых группах. Наоборот, чаще биопленки в той или иной степени формировали штаммы других филогрупп (р<0,05). Связь между биопленкообразованием и гемолитической активностью также не обнаружена (р>0,05).
Независимо от источника выделения штаммы уропатогенной E. coli в большинстве случаев были устойчивыми к ампициллину (62,1%). Зарегистрирован высокий уровень резистентности к ингибиторозащищенному пенициллину – амоксициллину/клавуланату (27,8% культур), цефотаксиму (37,9%) и ципрофлоксацину (36,9%). Среди нозокомиальных UPEC доля штаммов, устойчивых к амоксициллину/клавуланату и ципрофлоксацину, статистически значимо различалась между уринарными и катетер-ассоциированными культурами (р<0,05). В отношении всех UPEC наибольшей активностью обладали амикацин, фосфомицин и меропенем: доля чувствительных штаммов составила 90,9, 100 и 98,9% соответственно. Продукция БЛРС была выявлена у 51 (25,8%) культуры E. coli, при этом статистически значимо их доля различалась в группах нозокомиальных штаммов: уринарные<катетер-ассоциированные (р=0,0041). По количеству продуцирующих БЛРС E. coli в поликлинике и стационаре достоверных различий не выявлено, хотя в больничных условиях их удельный вес был все же выше. Интересно, что в поликлинике среди продуцентов БЛРС 17 (77,3%) штаммов были устойчивыми к ципрофлоксацину и 3 (13,6%) – к амикацину, а в стационаре таковых оказалось 25 (86,2%) и 5 (17,2%) соответственно, но разница между группами была также статистически не значимой. Связи между продукцией БЛРС и принадлежностью к филогруппе В2 не выявлено ни в отдельно сравниваемых группах поликлинических и нозокомиальных культур, ни в общей выборке, хотя В2-изоляты продуцировали БЛРС чаще, чем представители других филогрупп (28,9 против 23,4%).
Обсуждение. Ведущая этиологическая значимость бактерий E. coli в ИМВП обусловлена их доминированием среди факультативно-анаэробной микробиоты кишечного тракта, откуда они попадают в органы мочевыделительной системы восходящим путем [16, 17]. Распространенность ИМВП зависит от пола и возраста пациентов – встречаемость выше среди женщин и увеличивается с возрастом (кроме раннего младенчества). Согласно действующим европейским рекомендациям, диагностическим критерием этиологической значимости микроорганизма считается титр ≥103 КОЕ/мл, но микробное число различается в зависимости от нозологии и стадии заболевания [1]. Так, например, при остром обструктивном пиелонефрите показаны широкие диапазоны бактериурии для большинства микроорганизмов – от 101 до 106 КОЕ/мл [18]. Представленная в табл. 1 структура штаммов E. coli, выделенных от пациентов с различными ИМВП в рамках годичного исследования, позволяет считать данные выборки репрезентативными для изучения биологических свойств UPEC.
Большинство случаев ИМВП вызывается одним видом микроорганизма, и определение в образцах нескольких видов бактерий чаще всего объясняется нарушениями техники сбора и/или транспортировки материала. Однако у больных с аномалиями органов мочевыделительной системы, почечнокаменной болезнью, у пожилых людей с диабетом и др. при хроническом течении инфекции возможно выявление полимикробных ассоциаций [19–21]. Последние чаще детектируются при катетер-ассоциированных ИМВП в условиях стационара. Действительно, в настоящем исследовании в поликлинической группе доля ассоциаций составила около 20%, тогда как в стационарной – более 50%, а при катетеризации частота микст-вариантов выявлена уже в 80% исследований (см. табл. 2). При катетеризации двух-, трех- и четырехкомпонентные ассоциации встречались одинаково часто, что согласуется с данными других исследований, в которых показано, что при долгосрочной катетеризации обычно изолируют представителей 3–5 видов микроорганизмов [22]. Видовой спектр бактерий-ассоциантов существенно различался в двух исследуемых группах: в отличие от поликлинических E. coli, с которыми выделялись лишь энтеробактерии, энтерококки, коринебактерии и стафилококки, нозокомиальные штаммы изолированы в смешанной культуре с разнообразными микроорганизмами. Таксономический состав бактерий, выделенных вместе с E. coli при катетер-ассоциированных инфекциях, был ограничен лишь несколькими видами, что полностью согласуется с данными других исследований [5, 23]. Разнообразные коринебактерии, изолированные как у поликлинических, так и у пациентов в стационаре, вероятно, являются частью комменсальной микробиоты мочеполового тракта. Потенциальное биологическое и клиническое значение полимикробных ассоциаций в моче зависит от многих факторов. С одной стороны, обнаружение полимикробной ассоциации бактерий в моче является обоснованным поводом для подозрения на загрязнение образца периуретральной и/или вагинальной микробиотой, что действительно происходит в 20–25% случаев [20]. Кроме того, полимикробный характер инфекции чаще всего встречается среди людей с факторами риска развития ИМВП, поэтому большинство микроорганизмов-ассоциантов, являющихся членами «нормальной» микробиоты, могут быть упущены при интерпретации результатов бактериологического исследования. С другой стороны, существуют исследования по обнаружению идентичных полимикробных ассоциаций в моче и в крови, т.е. инфекция кровотока «поддерживает» интерпретацию полимикробного характера ИМВП [24]. Результаты метагеномного анализа также указывают на то, что при заболеваниях мочевой системы смешанные инфекции могут быть распространены значительно шире, чем принято считать [25].
Показано, что E. coli отличается значительной внутривидовой гетерогенностью и для штаммов определенных филогенетических групп характерны преимущественная локализация развития инфекционного процесса, а также патогенетические особенности их персистирования в организме [26]. В 2000 г. O. Clermont et al. разработали метод триплексной полимеразной цепной реакции (triplex ПЦР), согласно которому определенные гены или фрагменты ДНК могут быть специфическими маркерами филогенетических групп [27]. Впоследствии были предложены дополнительные филогенетические маркеры (quadriplex ПЦР), количество филогрупп расширено до восьми распознаваемых, семь из которых принадлежат к E. coli sensu stricto (A, B1, B2, C, D, E, F), а одна соответствует скрытой группе I ( cryptic clade I) [15]. Данных по филогенетическому разнообразию UPEC с учетом нового филогенетического анализа недостаточно, хотя реклассификация E. coli, изолированных из разных источников, идет довольно активно [28]. В кросс-секционном исследовании пациентов с ИМВП, принятых или госпитализированных в медицинских клиниках Ирана, продемонстрировано, что среди UPEC могут встречаться представители всех филогрупп [29]. Еще одно исследование проведено в Пакистане, где изоляты UPEC были распределены только в пяти филогруппах [12]. Оба исследования включали штаммы, собранные как в урологических отделениях, так и в амбулатории, и в обоих доминировала E. coli филогруппы В2, но доли других представителей существенно различались. Филогенетическое разнообразие внебольничных и внутрибольничных штаммов E. coli показано только в одном доступном нам исследовании, проведенном в Дании в 2012 г., но авторы изучали штаммы, изолированные из крови пациентов с ИМВП, и использовали ПЦР еще с тремя маркерными генами [30]. Сравнительный анализ по филогенетическому распределению поликлинических и нозокомиальных изолятов UPEC в клиниках г. Перми выявил, что первая группа была более разнообразной, тогда как в стационаре доминировали представители филогруппы В2 (см. табл. 3). Расчет отношения шансов (OR) показал, что встречаемость данных бактерий в стационаре в 12 раз выше, чем в поликлинике (OR=12,615). Интересно, что E. coli филогруппы D и F (ранее их относили к филогруппе D [31]) были детектированы только в 16 (7,6%) случаях, хотя это вторая по распространенности группа. Сравнение встречаемости этих представителей между нозокомиальными и амбулаторными штаммами не выявило статистически значимой разницы. E. coli филогруппы Е встречалась достоверно чаще в поликлинике.
Гемолитическая активность E. coli определяется продукцией четырех типов гемолизинов, но наиболее важным секретируемым фактором вирулентности UPEC является липопротеин, называемый α-гемолизином (HlyA). Его цитотоксический эффект обусловлен формированием пор в клетках уроэпителия, что ведет к их лизису и способствует бактериальной инвазии через эпителиальный барьер [2]. Показано, что встречаемость гемолитической E. coli существенно выше среди UPEC, чем среди фекальных симбиотических штаммов, и может достигать 30% [32]. Оценка гемолиза у штаммов UPEC, выделенных от пациентов поликлиник и стационаров, выявила, что каждый пятый штамм был гемолитическим (20,2%), но в стационаре гемолитическая E. coli филогруппы В2 встречается достоверно чаще.
Анализ биопленкообразующей способности штаммов UPEC показал, что поликлинические культуры не формировали биопленку в 24,8% случаев, стационарные – в 34,4%, при этом в последней группе небиопленкообразующими чаще были штаммы с катетеров, чем уринарные культуры. Связи между высоким биопленкообразованием и принадлежностью к филогруппе В2 не выявлено ни в общей, ни в отдельно сравниваемых группах, хотя в той или иной степени биопленки формировали более 60% культур филогруппы В2. В исследовании, проведенном на штаммах E. coli, изолированных от женщин с хронической или рецидивирующей ИМВП, также показано, что уровень их биопленкообразования in vitro был не выше, чем в группе других UPEC [33].
Анализ антибиотикочувствительности возбудителей ИМВП регулярно проводится за рубежом и в России. Обсуждаемой проблеме посвящено несколько многоцентровых исследований (UTIAP-1, -2, -3; АРИМБ, АРМИД, ДАРМИС) в рамках работы научно-методического центра Минздрава России по мониторингу антибиотикорезистентности, созданного на базе НИИ антимикробной химиотерапии СГМА. В последнем многоцентровом исследовании антибиотикорезистентности возбудителей внебольничных ИМВП, проведенном в 2010–11 гг., выявлено, что для взрослых характерна высокая частота выделения резистентных штаммов E. coli к ампициллину (46,6%), пиперациллину (42,2%), ингибиторозащищенным пенициллинам – ампициллину/сульбактаму (40,1%) и амоксициллину/клавуланату (41,7%), ципрофлоксацину (20,1%). Отмечено, что частота встречаемости БЛРС-продуцирующих штаммов E. coli, выделенных от амбулаторных больных, составила 8,5%, хотя в 2005 г. была равной 2% [34]. В Перми доли поликлинических штаммов UPEC, устойчивых к ампициллину и ципрофлоксацину, а также БЛРС-продуцентов оказались выше – 66,7% (на 20%), 34,3% (на 14%) и 21% (в 2,5 раза) соответственно, т.е. тенденции к снижению чувствительности возбудителей внебольничных ИМВП к данным антибиотикам и к увеличению распространенности БЛРС-изолятов, выявленные несколько лет назад, сохраняются. Следует отметить, что большой процент БЛРС-продуцирующих штаммов UPEC показан и в других исследованиях, например, в Пакистане выделено 43,2% изолятов от пациентов с ИМВП [35].
Проведенный в Словении анализ антибиотикоустойчивости штаммов UPEC не выявил связи между наличием множественной лекарственной устойчивости (MDR) и принадлежностью изолята к филогруппе B2 [36]. Позднее D. Iranpour et al. [29], наоборот, продемонстрировали, что MDR-изоляты в половине случаев принадлежали к филогруппе B2. Millán Y. et al. (2014) обнаружили высокий процент представителей филогруппы В2 среди изолятов UPEC, продуцирующих β-лактамазу CTX-M-15 [10]. В данном исследовании связи между продукцией БЛРС и принадлежностью к филогруппе В2 не выявлено, но среди продуцирующих бета-лактамазы UPEC, доля В2-представителей была наибольшей. Увеличение распространенности UPEC филогруппы В2, устойчивых к фторхинолонам и продуцирующих БЛРС, представляет серьезную проблему, потому что штаммы этой группы, как известно, проявляют наибольшую вирулентность. Так, например, ИМВП-ассоциированный, полиантибиотикорезистентный штамм E. coli ST131, продуцирующий многочисленные факторы вирулентности, встречается во всем мире [37].
Заключение. Изучены биологические свойства и филогенетическое разнообразие UPEC, выделенных при ИМВП у амбулаторных и стационарных больных. Показано, что поликлинические и нозокомиальные изоляты существенно различались по встречаемости представителей различных филогрупп. В поликлинике детектированы E. coli всех восьми распознаваемых групп, тогда как в стационаре доминировали представители филогруппы В2, к которой отнесены все катетер-ассоциированные штаммы. Поликлинические UPEC в смешанной культуре выделялись только в 20%, тогда как нозокомиальные – более чем в 50% случаев, а при катетеризации частота микст-вариантов составила 80%, но с ограниченным видовым спектром бактерий-ассоциантов. Гемолитическая и биопленкообразующая способности штаммов UPEC не различались в двух группах, но в той или иной степени биопленки формировали более 60% культур филогруппы В2, кроме того, в стационаре гемолитические E. coli филогруппы В2 встречались достоверно чаще, чем в поликлинике. Необходимо отметить снижение чувствительности внебольничных UPEC к ципрофлоксацину и бета-лактамным антибиотикам (за исключением карбапенемов), в том числе за счет продукции БЛРС, что способствует сближению фенотипов резистентности поликлинических и стационарных культур. Полученные результаты, с одной стороны, свидетельствуют, что принадлежность E. coli к определенной филогенетической группе не является определяющей при возникновении ИМВП, с другой – указывают на возможность концентрирования в стационаре в условиях иммунокомпрометированного хозяина представителей филогруппы В2 с высоким вирулентным потенциалом.



